17 Cytoplasmic Membranes
Learning Objectives
- Emphasize the dynamic nature of the endomembrane system within the cell.
- Elucidate the structure and function of the rough and smooth endoplasmic reticulum.
- Describe the basic synthetic pathways for secretory and integral membrane proteins.
- Describe the role and sites of glycosylation in the processing of secretory and integral membrane proteins.
- Elucidate the structure, function, and polarization of the Golgi complex.
- Explain the signals used to target proteins to their appropriate cellular location.
- Describe lysosomal structure and function and the diseases caused by lysosome malfunction.
- Distinguish between phagocytosis, bulk phase endocytosis, and receptor-mediated endocytosis.
17.1 Cellular Organelles and the Endomembrane System
Organization of the nucleus
Like most other cellular organelles, the nucleus is surrounded by a membrane called the nuclear envelope. This membranous covering consists of two adjacent lipid bilayers with a thin fluid space in between them. Spanning these two bilayers are nuclear pores. A nuclear pore is a tiny passageway for the passage of proteins, RNA, and solutes between the nucleus and the cytoplasm.
Proteins called pore complexes lining the nuclear pores regulate the passage of materials into and out of the nucleus.
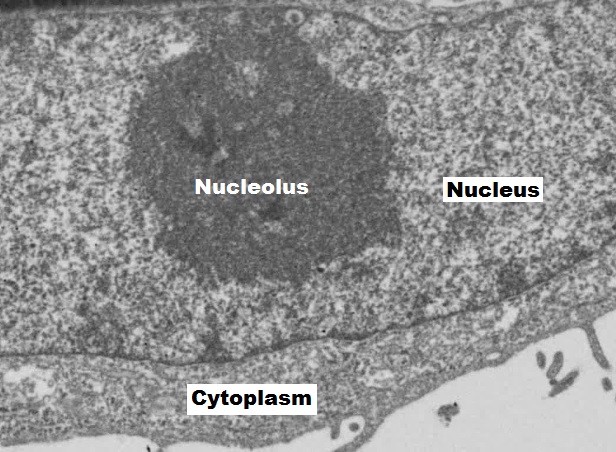
Inside the nuclear envelope is a gel-like nucleoplasm with solutes that include the building blocks of nucleic acids. There also can be a dark-staining mass often visible under a simple light microscope, called a nucleolus (plural = nucleoli). The nucleolus is a region of the nucleus that is responsible for manufacturing the RNA necessary for construction of ribosomes. Once synthesized, newly made ribosomal subunits exit the cellʼs nucleus through the nuclear pores (figure 17.1). Proteins entering the nucleus require nuclear localization signals, while proteins exiting require nuclear export signals.
Endomembrane system
The endomembrane system (endo = “within”) is a group of membranes and organelles (figure 17.2) in eukaryotic cells that work together to modify, package, and transport lipids and proteins. It includes the nuclear envelope as well as:
- Endoplasmic reticulum,
- Golgi apparatus,
- Endosomes,
- Lysosomes,
- Vacuoles, and
- Peroxisomes.
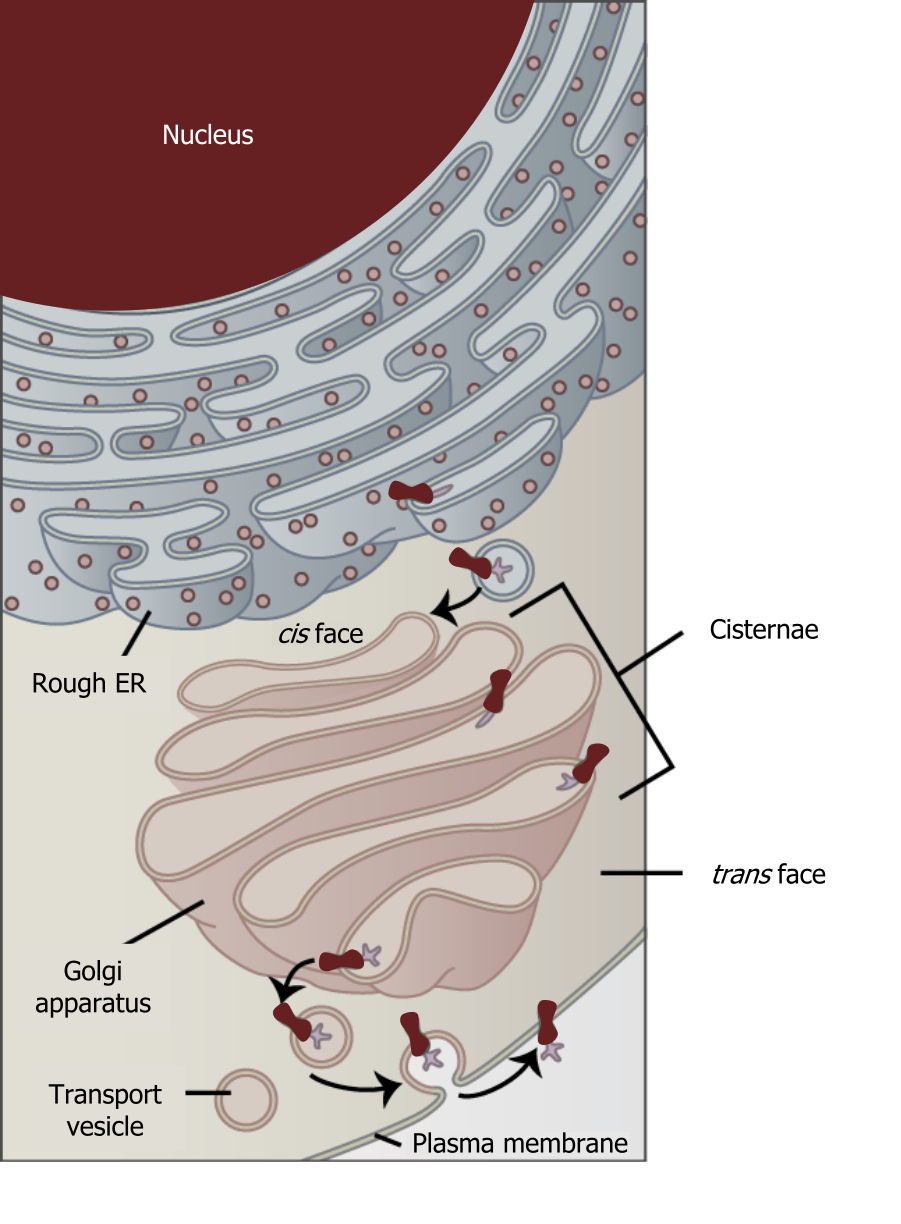
Although not technically within the cell the plasma membrane is included in the endomembrane system because, it interacts with the other endomembranous organelles. The endomembrane system does not include the mitochondria. The system of intracellular membranes is designed to move proteins through both the secretory pathway (constitutive or regulated) and the endocytic pathways.
The endoplasmic reticulum (ER)
The endoplasmic reticulum (ER) (figure 17.2) is a series of interconnected membranous sacs and tubules that collectively modify proteins and synthesize lipids. However, these two functions take place in separate areas of the ER: the rough ER and the smooth ER, respectively.
Smooth ER
The smooth endoplasmic reticulum (SER) is continuous with the rough ER (RER) but has few or no ribosomes on its cytoplasmic surface. SER functions include synthesis of carbohydrates, lipids, and steroid hormones; detoxification of medications and poisons; and storing calcium ions. In muscle cells, a specialized SER, the sarcoplasmic reticulum, is responsible for storing calcium ions that are needed to trigger the muscle cellsʼ coordinated contractions.
Rough ER
Scientists have named the rough endoplasmic reticulum (RER) as such because the ribosomes attached to its cytoplasmic surface give it a studded appearance when viewing it through an electron microscope.
Ribosomes transfer their newly synthesized proteins into the RERʼs lumen where they undergo structural modifications, such as folding or acquiring side chains. These modified proteins incorporate into cellular membranes, the ER, or other organellesʼ membranes. The proteins can also be secreted from the cell (such as protein hormones and enzymes). The RER also makes phospholipids for cellular membranes.
If the phospholipids or modified proteins are not destined to stay in the RER, they will reach their destinations via transport vesicles that bud from the RERʼs membrane.
Glycosylation
Nearly all RER-synthesized proteins are glycosylated with short-branched oligosaccharides. This occurs in an N-linked fashion on asparagine residues.
Protein degradation
If proteins arenʼt folded properly, this can contribute to a host of disease processes related to misfolding events. Typically, folding is facilitated in the ER using chaperones (BiP), but if the protein is altered (due to mutation), this can lead to aggregation. Accumulation of BiP can initiate the unfolded protein response (UPR) (figure 17.3).
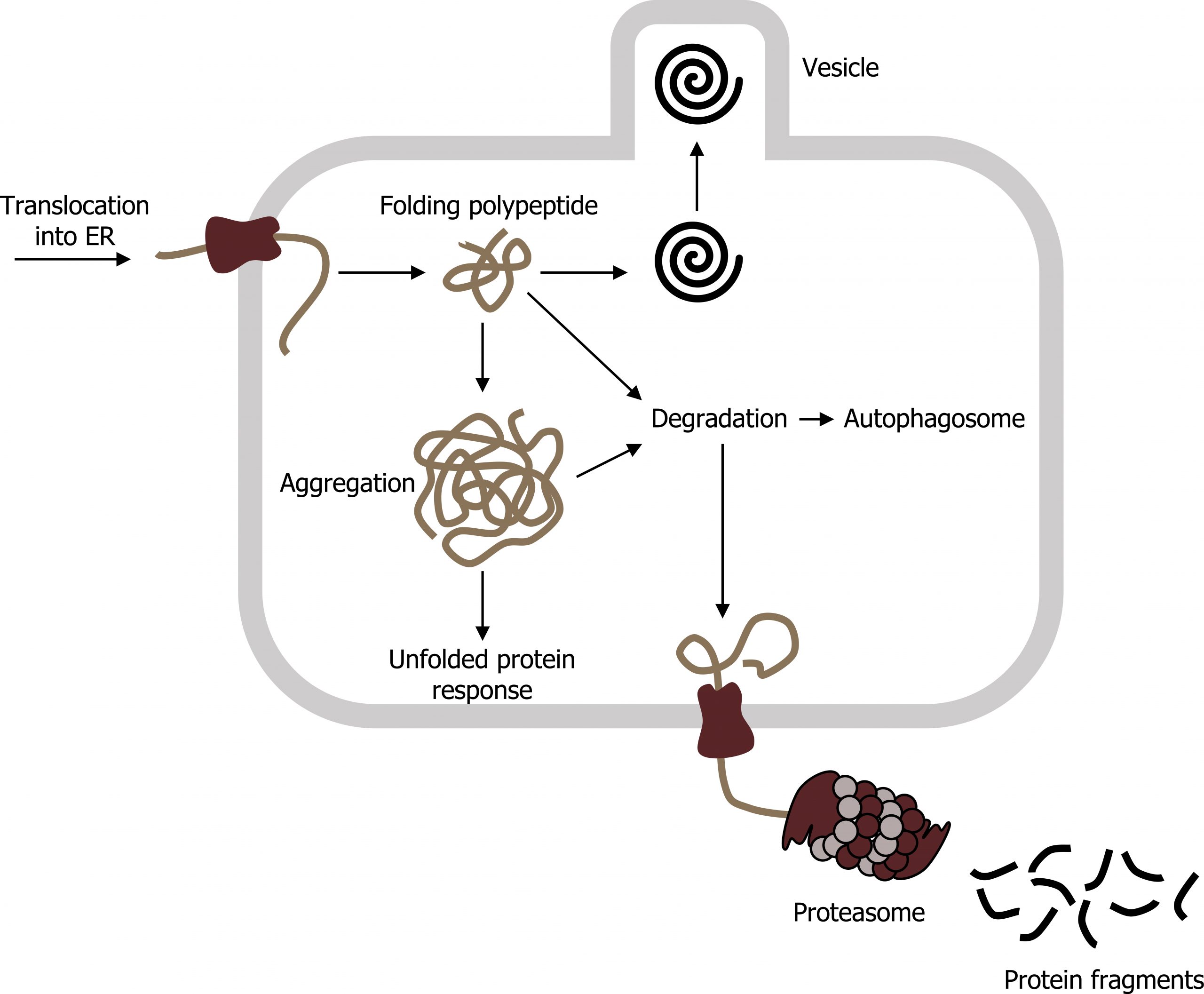
E3 ubiquitin ligase is often responsible for tagging aggregates with ubiquitin, which targets the protein to the proteasome. The proteasome consists of two subunits (19S and 20S) to make a functional 26S proteasome. Inside the proteasome, the polypeptide chains are cleaved back to their native amino acids and can be reused in other translational events. However, if the aggregates accumulate, in some instances they can contribute to any number of neurodegenerative disorders.
Golgi
When proteins exit the RER they are trafficked to the Golgi where they will incur further post-translational modifications and will translocate to their final destination. These modifications will include “pruning” of large oligosaccharides that were attached in the RER, glycosylation, sulfation, and phosphorylation. Additionally, some proteins require Golgi-associated cleavage to produce a mature protein ready for trafficking.
The Golgi is divided into trans and cis networks.
- Cis Golgi is close to the ER and sorts proteins back to the ER.
- Trans Golgi sorts proteins into vesicles bound for the plasma membrane and intracellular vesicles.
In the Golgi, O-linked glycosylation happens, and most mannose residues are removed. This is done by a large family of enzymes known as glycosyltranferases.
Protein localization
Protein translation can take place on both free ribosomes and the RER. Free ribosomes translate proteins bound for the mitochondria, nucleus, and peroxisomes.
The RER translates proteins for secretion, membrane-bound proteins, or soluble proteins.
Proteins translated on the RER are folded and processed into mature proteins in the lumen of the ER. Transcripts for protein products to be translated on the RER are characteristic of a signal sequence that is recognized by a signal recognition peptide. The signal sequence on the nascent polypeptide will be used to later target the protein to its correct location. The signal recognition peptide facilitates the docking of the ribosome complex on the ER, and the peptide is translated into the lumen of the ER. Inside the ER, the peptide is often associated with chaperones to assist in correct protein folding.
Localization using coat proteins (COP)
Once translated in the RER, proteins are trafficked in the cell using vesicle transport systems. The direction of the transport, ER to Golgi or Golgi to ER, is determined by the coat proteins on the vesicles.
- Retrograde transport: The coat protein COPI is used for retrograde transport, from the cis-Golgi to the ER.
- Anterograde transport: The coat protein COPII is used for anterograde transport from the ER to the cis-Golgi.
- Additional transport mechanisms: Clathrin is used for trans-Golgi transport to lysosomes. It is also used in endocytosis in transporting proteins from the plasma membrane to the endosomes.
The vesicles are targeted to the intended membrane by transport over microtubules in the cytosol. The fusion itself requires surface proteins, Snares, which facilitate the formation of a docking complex stabilizing the interaction between the vesicle and the intended membrane. GTP is required for fusion of the two membranes.
Lysosomes and peroxisomes
Lysosomes are organelles formed by the fusion of a late endosome and a lysosomal-enzyme-filled vesicle secreted from the Golgi. Proteins are targeted to lysosomes by the presence of mannose 6-phosphate (acquired in the RER), and the presence of these tags are essential for trafficking to the lysosome.
The major function for these organelles is to break down macromolecules through enzymatic degradation. Both processes of autophagy and exocytosis can be facilitated. Lysosomal storage diseases are inherited metabolic diseases characterized by an abnormal buildup of various metabolic intermediates. Collectively, there are approximately fifty of these disorders, and they may affect different parts of the body. Clinical correlates include: Gaucher disease, Fabry disease, glycogen storage disease, mucopolisacaridosis, and sphingolipidoses.
This is in contrast to peroxisomes, which are formed by budding from the ER. They primarily perform hydrogen peroxide-mediated degradation of lipids (i.e., very long-chain fatty acids) and some amino acids. Zellweger syndrome is one of the heritable disorders of peroxisome biogenesis and results in infant death before six months.
17.1 References and resources
Text
Clark, M. A. Biology, 2nd ed. Houston, TX: OpenStax College, Rice University, 2018, Chapter 4: Cell Structure, Chapter 5: Structure and Function of the Plasma Membranes.
Karp, G., and J. G. Patton. Cell and Molecular Biology: Concepts and Experiments, 7th ed. Hoboken, NJ: John Wiley, 2013, Chapter 8: Cytoplasmic Membrane Systems: Structure, Function, and Membrane Trafficking.
Le, T., and V. Bhushan. First Aid for the USMLE Step 1, 29th ed. New York: McGraw Hill Education, 2018, 46–47.
Figures
Grey, Kindred, Figure 17.2 Interaction of the endomembrane systems. 2021. https://archive.org/details/17.2_20210926. CC BY 4.0. Adapted from Figure 4.18. CC BY 4.0. From OpenStax.
Grey, Kindred, Figure 17.3: Unfolded protein response in the RER. 2021. CC BY 4.0. Adapted from ProteinQS en by Vojtěch Dostál. Public domain. From Wikimedia Commons.
Orlov I, Schertel A, Zuber G, et al. Figure 17.1 EM of the nucleus and nucleolus. CC BY-SA 4.0. From Wikimedia Commons.
Additional resources
- Some helpful animations related to this content
- Translocation into ER: https://youtu.be/pxG8-BWbpnU
- Proteasome: https://youtu.be/Gp8lhKghckY
- Clathrin: https://youtu.be/o_EUHu4OJus
17.2 Endocytosis
Endocytosis
Endocytosis is a type of active transport that moves particles, such as large molecules, parts of cells, and even whole cells, into a cell. There are different endocytosis variations, but all share common characteristics:
- The cellʼs plasma membrane invaginates, forming a pocket around the target particle.
- The pocket pinches off.
- This results in the particle containing itself in a newly created intracellular vesicle formed from the plasma membrane.
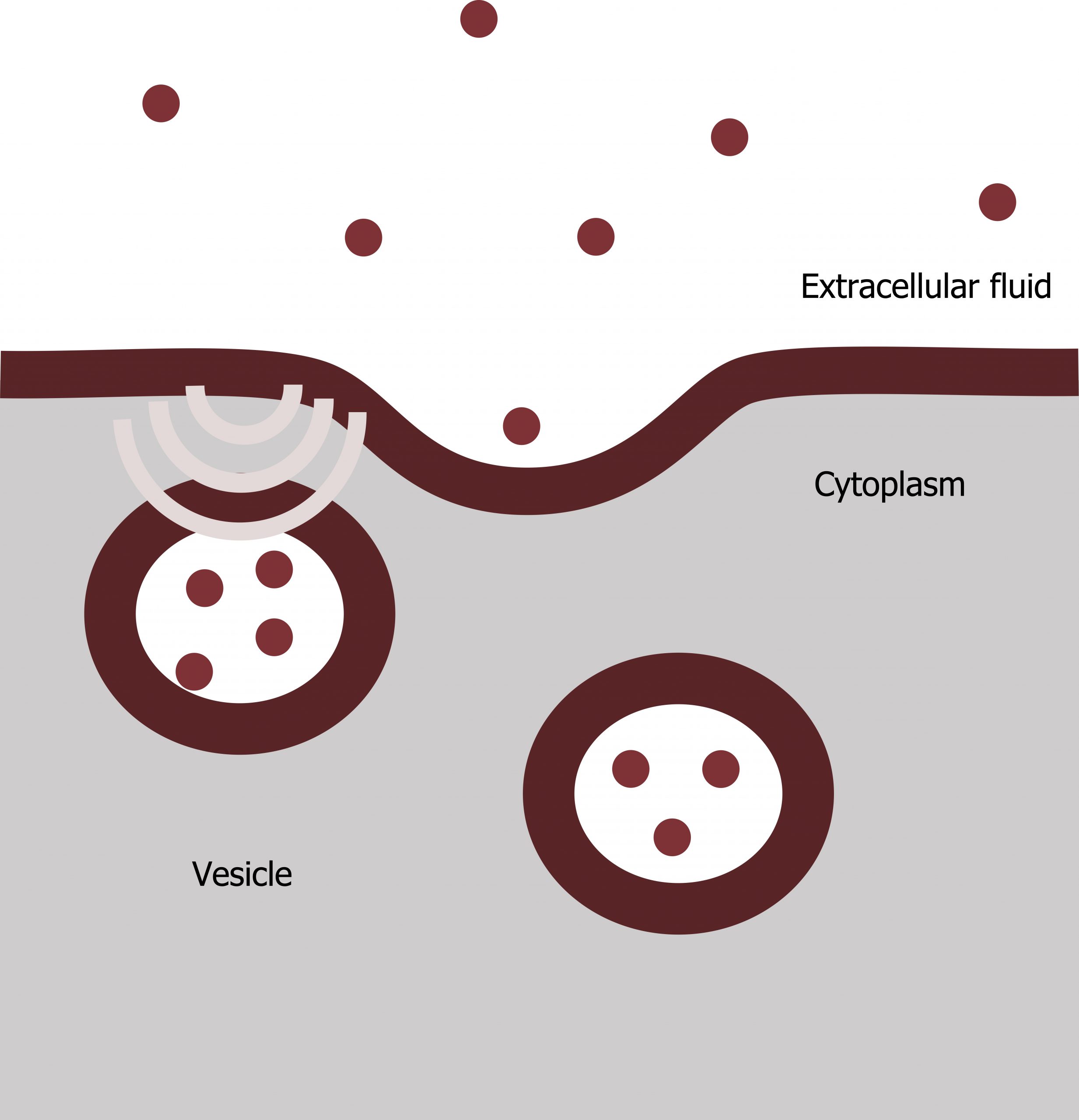
Phagocytosis
Phagocytosis (the condition of “cell eating”) is the process by which a cell takes in large particles, such as other cells or relatively large particles. For example, when microorganisms invade the human body, a type of white blood cell, a neutrophil, will remove the invaders through this process, surrounding and engulfing the microorganism, which the neutrophil then destroys (figure 17.4).
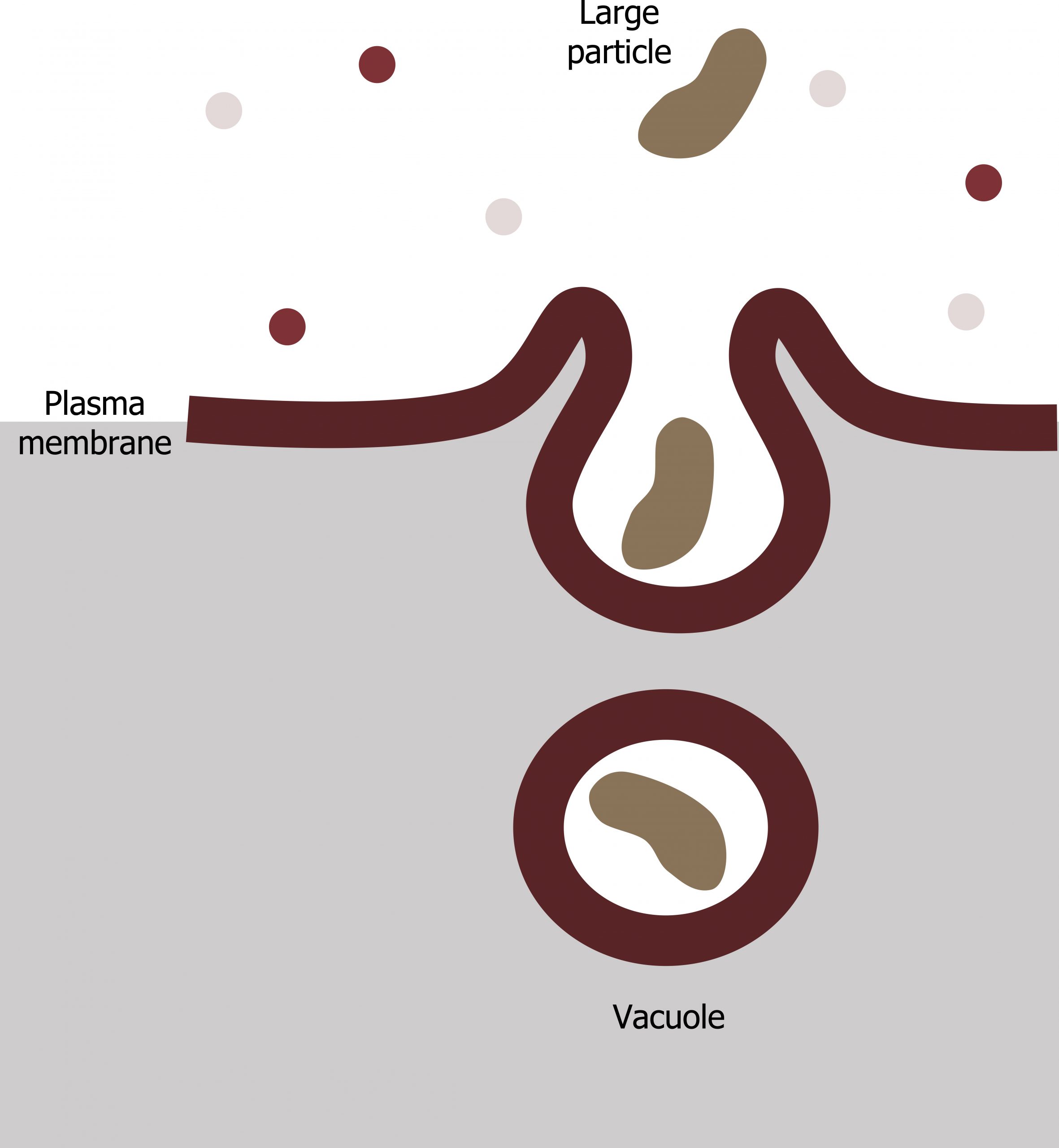
In preparation for phagocytosis, a portion of the plasma membraneʼs inward-facing surface becomes coated with the protein clathrin, which stabilizes this membraneʼs section. The membraneʼs coated portion then extends from the cellʼs body and surrounds the particle, eventually enclosing it. Once the vesicle containing the particle is enclosed within the cell, the clathrin disengages from the membrane ,and the vesicle merges with a lysosome for breaking down the material in the newly formed compartment (endosome). When accessible nutrients from the vesicular contentsʼ degradation have been extracted, the newly formed endosome merges with the plasma membrane and releases its contents into the extracellular fluid. The endosomal membrane again becomes part of the plasma membrane.
Receptor-mediated endocytosis
A targeted variation of endocytosis employs receptor proteins in the plasma membrane that have a specific binding affinity for certain substances (figure 17.5).
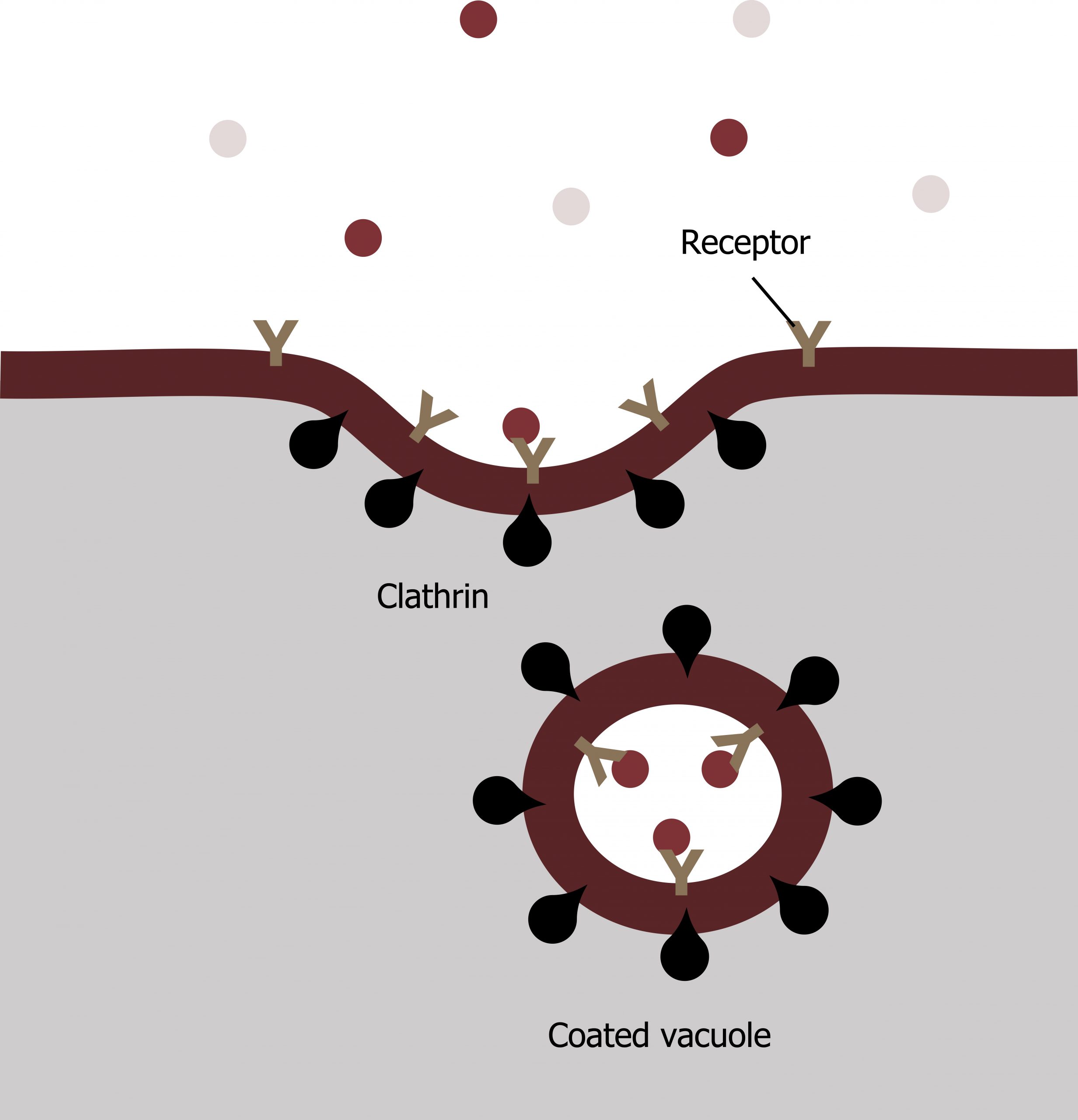
In receptor-mediated endocytosis, the cellʼs uptake of substances targets a single type of substance that binds to the receptor on the cell membraneʼs external surface.
Clathrin attaches to the plasma membraneʼs cytoplasmic side. If a compoundʼs uptake is dependent on receptor-mediated endocytosis and the process is ineffective, the material will not be removed from the tissue fluids or blood. Instead, it will stay in those fluids and increase in concentration. The failure of receptor-mediated endocytosis causes some human diseases.
For example, receptor-mediated endocytosis removes low-density lipoprotein or LDL from the blood. In the human genetic disease familial hypercholesterolemia, the LDL receptors are defective or missing entirely. People with this condition have life-threatening levels of cholesterol in their blood because their cells cannot clear LDL particles. See chapter 6.
Exocytosis
Exocytosis is the opposite of the processes we discussed above in that its purpose is to expel material from the cell into the extracellular fluid. Waste material is enveloped in a membrane and fuses with the plasma membraneʼs interior. This fusion opens the membranous envelope on the cellʼs exterior, and the waste material expels into the extracellular space. Other examples of cells releasing molecules via exocytosis include extracellular matrix protein secretion and neurotransmitter secretion into the synaptic cleft by synaptic vesicles (figure 17.6).
17.2 References and resources
Text
Clark, M. A. Biology, 2nd ed. Houston, TX: OpenStax College, Rice University, 2018, Chapter 4: Cell Structure, Chapter 5: Structure and Function of the Plasma Membranes.
Karp, G., and J. G. Patton. Cell and Molecular Biology: Concepts and Experiments, 7th ed. Hoboken, NJ: John Wiley, 2013, Chapter 8: Cytoplasmic Membrane Systems: Structure, Function, and Membrane Trafficking.
Le, T., and V. Bhushan. First Aid for the USMLE Step 1, 29th ed. New York: McGraw Hill Education, 2018, 46–47.
Figures
Alberts B, Johnson A, Lewis J, et al. Figure 17.4 General process of phagocytosis… Adapted under Fair Use from Cell and Molecular Biology. 6th Ed. pp 308. Figure 8.46 A summary of phagocytic pathway. 2014.
Alberts B, Johnson A, Lewis J, et al. Figure 17.5 Receptor mediated endocytosis, LDL-receptor is a classic example of this process. Adapted under Fair Use from Cell and Molecular Biology. 6th Ed. pp 306. Figure 8.42 The endocytic pathway. 2014.
Alberts B, Johnson A, Lewis J, et al. Figure 17.6 Exocytosis: vesicles containing substances fuse with the plasma membrane… Adapted under Fair Use from Cell and Molecular Biology. 6th Ed. pp 299. Figure 35 A summary of the autophagic pathway. 2014.
Additional resources
- Some helpful animations related to this content
- Translocation into ER: https://youtu.be/pxG8-BWbpnU
- Proteasome: https://youtu.be/Gp8lhKghckY
- Clathrin: https://youtu.be/o_EUHu4OJus

