10 Genes, Genomes, and DNA
Learning Objectives
- Define the word genome, and describe what composes the eukaryotic genome.
- Describe the chemical structure and physical properties of DNA.
- Outline the levels of structure of chromatin and chromosomes.
- Define the types of chromatin (heterochromatin, euchromatin, and facultative and constitutive chromatin).
- Describe the characteristics of nonrepetitive DNA, highly repetitive DNA, and moderately repetitive DNA and its relative abundance and significance throughout the genome.
- Describe sources and types of genetic polymorphism.
- Describe the mechanisms and implications of different classes of genetic mutation (genome mutation, chromosome mutation, and gene mutation).
- Emphasize the bidirectional and semidiscontinuous nature of DNA replication and the reasons for it.
- Outline the different types of DNA damage caused by various environmental conditions.
- Outline what is known about the different types of DNA repair mechanisms.
About this Chapter
The cell is the most fundamental unit of all eukaryotic organisms. Its components and their cellular interactions are essential to the inner workings of the cell as well as influencing the surrounding environment through cell interactions. Starting at the level of DNA, we will address how individual nucleotide changes can alter cellular process leading to the change of a cell (and therefore organismal) phenotype.
10.1 DNA Structure
Nucleotides and basic DNA structure
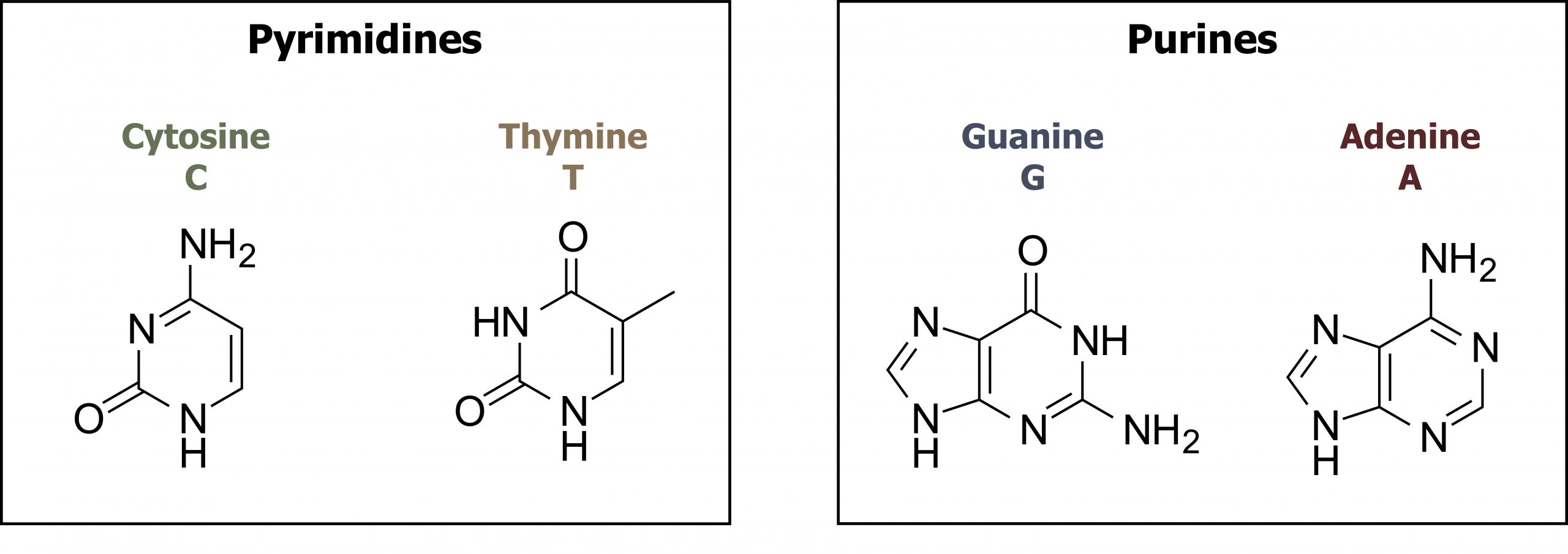
The building blocks of DNA are nucleotides. The important components of the nucleotide are a nitrogenous (nitrogen-bearing) base, a five-carbon sugar (pentose), and a phosphate group. The nucleotide is named depending on the nitrogenous base. The nitrogenous base can be a purine, such as adenine (A) and guanine (G), or a pyrimidine, such as cytosine (C) and thymine (T). The purines have a double-ring structure with a six-membered ring fused to a five-membered ring. Pyrimidines are smaller in size; they have a single six-membered ring structure. The sugar is deoxyribose in DNA and ribose in RNA. The carbon atoms of the five-carbon sugar are numbered 1′, 2′, 3′, 4′, and 5′ (1′ is read as “one prime”). The phosphate, which makes DNA and RNA acidic, is connected to the 5′ carbon of the sugar by the formation of an ester linkage between phosphoric acid and the 5′-OH group (an ester is an acid + an alcohol). In DNA nucleotides, the 3′ carbon of the sugar deoxyribose is attached to a hydroxyl (OH) group (figures 10.1 and 10.2).
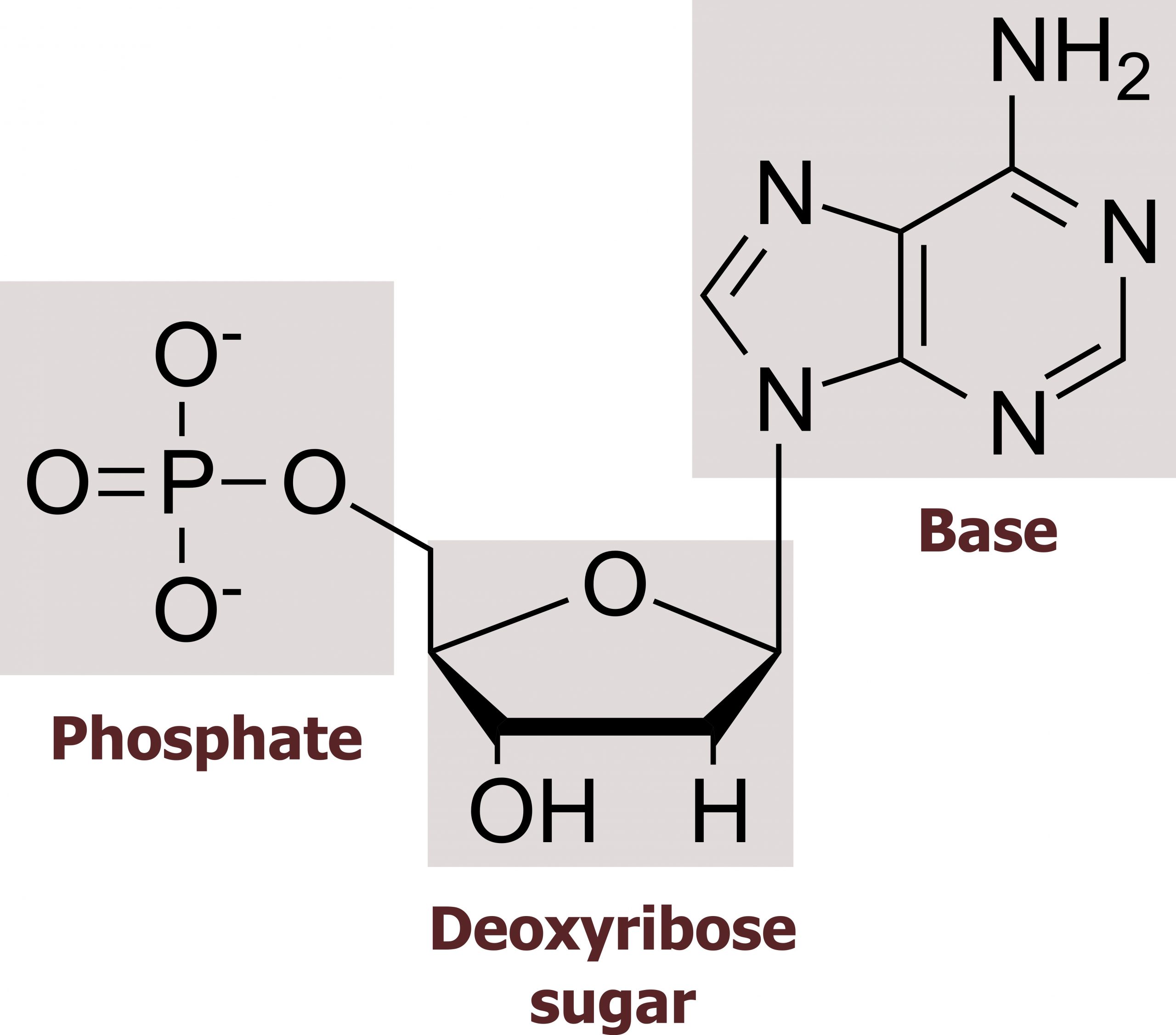
The nucleotides combine with each other to produce phosphodiester bonds. The phosphate residue attached to the 5′ carbon of the sugar of one nucleotide forms a second ester linkage with the hydroxyl group of the 3′ carbon of the sugar of the next nucleotide, thereby forming a 5′-3′ phosphodiester bond. In a polynucleotide, one end of the chain has a free 5′ phosphate, and the other end has a free 3′-OH. These are called the 5′ and 3′ ends of the chain.
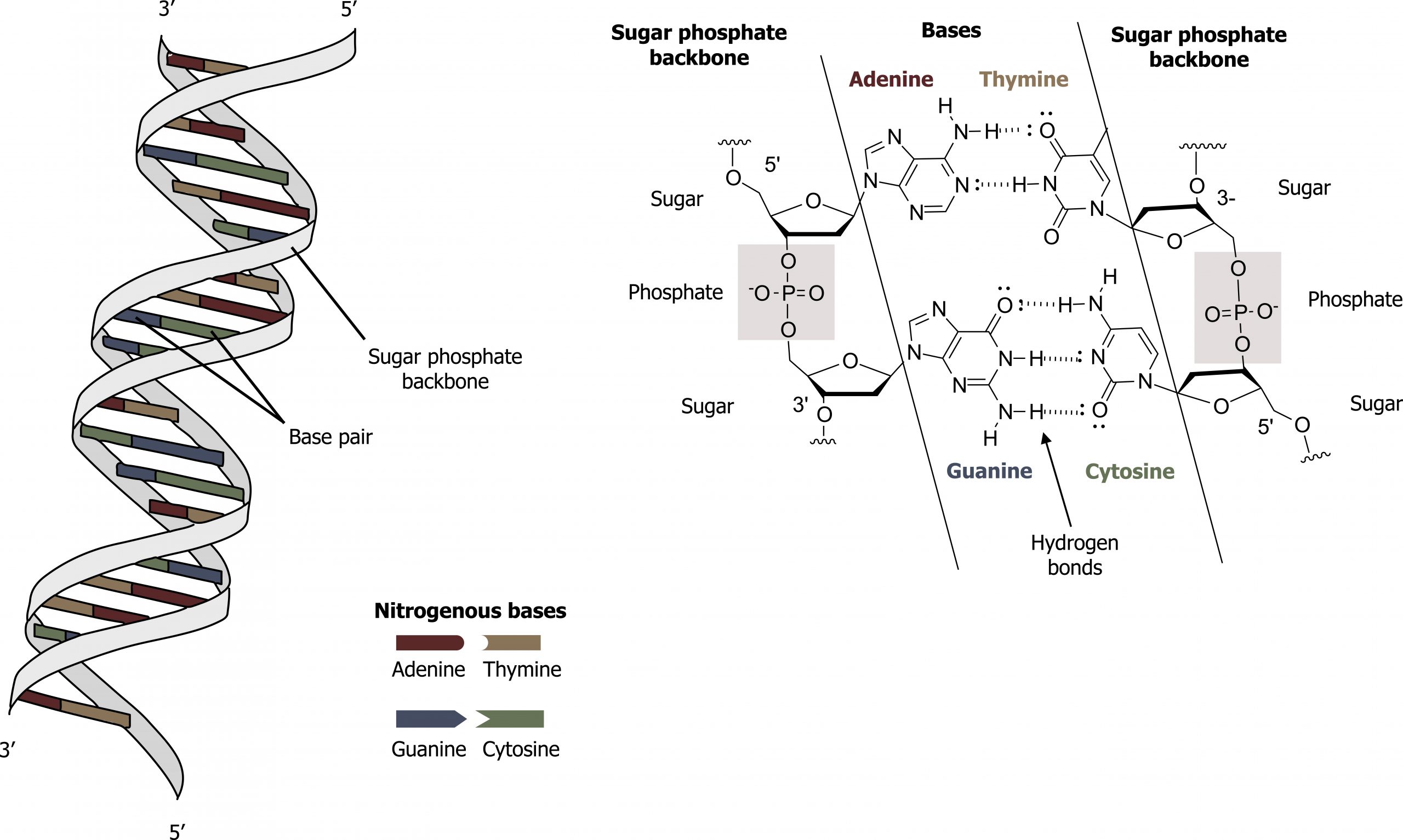
Base-pairing takes place between a purine and pyrimidine on opposite strands, so that adenine and thymine are complementary base pairs, and cytosine and guanine are also complementary base pairs. The base pairs are stabilized by hydrogen bonds: adenine and thymine form two hydrogen bonds, and cytosine and guanine form three hydrogen bonds. The two strands are anti-parallel in nature; that is, the 3′ end of one strand faces the 5′ end of the other strand. The sugar and phosphate of the nucleotides form the backbone of the structure, whereas the nitrogenous bases are stacked inside, like the rungs of a ladder. The twisting of the two strands around each other results in the formation of uniformly spaced major and minor grooves.
DNA has a double helix structure and phosphodiester bonds; the dotted lines between thymine and adenine and guanine and cytosine represent hydrogen bonds. The major and minor grooves are binding sites for DNA-binding proteins during processes such as transcription (the copying of RNA from DNA) and replication (figure 10.3).
DNA packaging and organization
Eukaryotic chromosomes consist of a linear DNA molecule complexed with protein (histones); this complex is called chromatin. Histones are evolutionarily conserved proteins that are rich in basic amino acids and form an octamer composed of two molecules of each of four different histones.
The DNA (remember, it is negatively charged because of the phosphate groups) is wrapped tightly around the histone core. This interaction is facilitated through electrostatic interactions. The negatively charged phosphate groups on the DNA backbone are attracted to a positively charged lysine on the exposed surface of histones. This nucleosome is linked to the next one with the help of a linker DNA. This is also known as the “beads on a string” structure. With the help of a fifth histone, a string of nucleosomes is further compacted into a 30 nm fiber, which is the diameter of the structure. Metaphase chromosomes are even further condensed by association with scaffolding proteins. At the metaphase stage, the chromosomes are at their most compact, approximately 700 nm in width (figure 10.4).
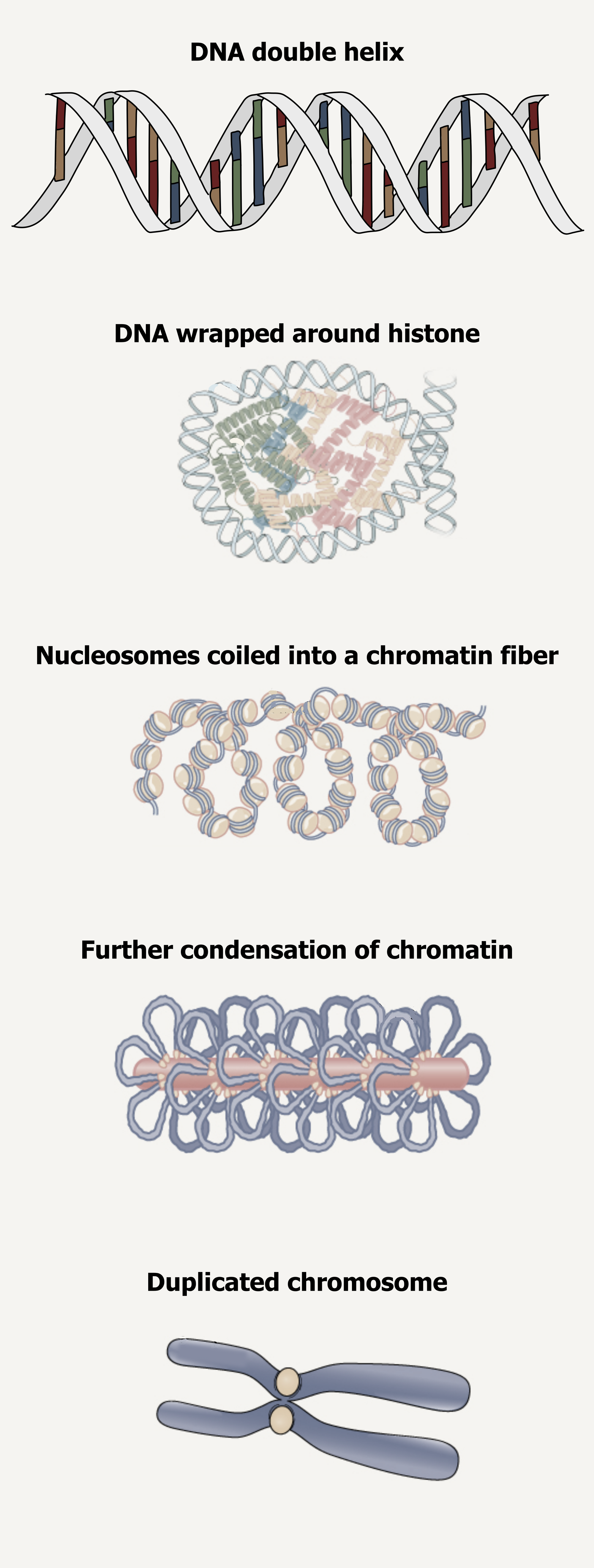
In interphase, eukaryotic chromosomes have two distinct regions that can be distinguished by staining. The tightly packaged region is known as heterochromatin, and the less dense region is known as euchromatin.
Heterochromatin usually contains genes that are not expressed and is found in the regions of the centromere and telomeres.
The euchromatin usually contains genes that are transcribed, with DNA packaged around nucleosomes but not further compacted.
Histone tails can be modified through both methylation and acetylation, which will alter the histone:DNA interaction. Histone methylation can have variable impacts on a given gene locus leading to a change in transcription. Histone acetylation relaxes the interactions of histones and DNA by removing the positive charge on lysine residues allowing the DNA to be transcriptionally accessible (euchromatin). DNA methylation, specifically to CpG islands, globally represses transcription. These modifications on histones and DNA can result in epigenetic influences that have an impact on many biological processes.
Across the three billion base pair genome, genes are organized into clusters with only a fraction of the DNA coding for translated products. The remaining DNA was historically considered “junk,” however, more recently there is a new appreciation for the roles of noncoding DNA regions. Only half of the genome is unique DNA sequence, and only 1.5 percent codes for mRNA (~20,000 protein-coding genes). The remaining sequence can be categorized as:
- Moderately repetitive: DNA containing ribosomal RNA (rRNA), tandem and nontandem repeats, and short and long interspersed nuclear elements (SINE and LINE).
- Transposable elements: These are movable elements, transposons or retrotransposons, that can result in disease-causing mutations if inserted into important genomic loci.
- Highly repetitive sequence: Satellites and mini satellites are regions of high sequence repetition (trinucleotide repeats) and are difficult to replicate. This can lead to expansions of these areas as well as mutations resulting in frame shifts or loss of translational starts.
10.1 References and resources
Text
Clark, M. A. Biology, 2nd ed. Houston, TX: OpenStax College, Rice University, 2018, Chapter 14: DNA Structure and Function.
Karp, G., and J. G. Patton. Cell and Molecular Biology: Concepts and Experiments, 7th ed. Hoboken, NJ: John Wiley, 2013, Chapter 10: The Nature of the Gene and the Genome, Chapter 12: The Cell Nucleus and the Control of Gene Expression, Chapter 13: DNA Replication.
Le, T., and V. Bhushan. First Aid for the USMLE Step 1, 29th ed. New York: McGraw Hill Education, 2018, 34, 38–40.
Nussbaum, R. L., R. R. McInnes, H. F. Willard, A. Hamosh, and M. W. Thompson. Thompson & Thompson Genetics in Medicine, 8th ed. Philadelphia: Saunders/Elsevier, 2016, Chapter 2: The Introduction to the Human Genome.
Figures
Grey, Kindred, Figure 10.1 Basic structure of nucleosides including the sugar (ribose or deoxyribose), base (pyrimidine or purine) and phosphate groups. 2021. Chemical structure by Henry Jakubowski. https://archive.org/details/10.1_20210926. CC BY 4.0.
Grey, Kindred, Figure 10.2 Structure of pyrimidine and purine bases. 2021. Chemical structure by Henry Jakubowski. https://archive.org/details/10.2_20210926. CC BY 4.0.
Grey, Kindred, Figure 10.3 General structure and hydrogen bonding pattern of DNA. 2021. Chemical structure by Henry Jakubowski. https://archive.org/details/10.3_20210926. CC BY-SA 4.0. Added DNA double helix grooves by Biochemlife. CC BY-SA 4.0. From Wikimedia Commons.
Grey, Kindred, Figure 10.4 Organizational structure of DNA illustrating condensation and supercoiling into chromosomes. 2021. https://archive.org/details/10.4_20210926. CC BY-SA 4.0. Added DNA double helix grooves by Biochemlife. CC BY-SA 4.0. From Wikimedia Commons. And Figure 14.11. CC BY 4.0. From OpenStax.
10.2 DNA Repair
DNA replication is a highly accurate process, but mistakes can occasionally occur, such as a DNA polymerase (DNA pol) inserting a wrong base. Uncorrected mistakes may sometimes lead to serious consequences, such as cancer. Repair mechanisms correct the mistakes. In rare cases, mistakes are not corrected, leading to mutations; in other cases, repair enzymes are themselves mutated or defective.
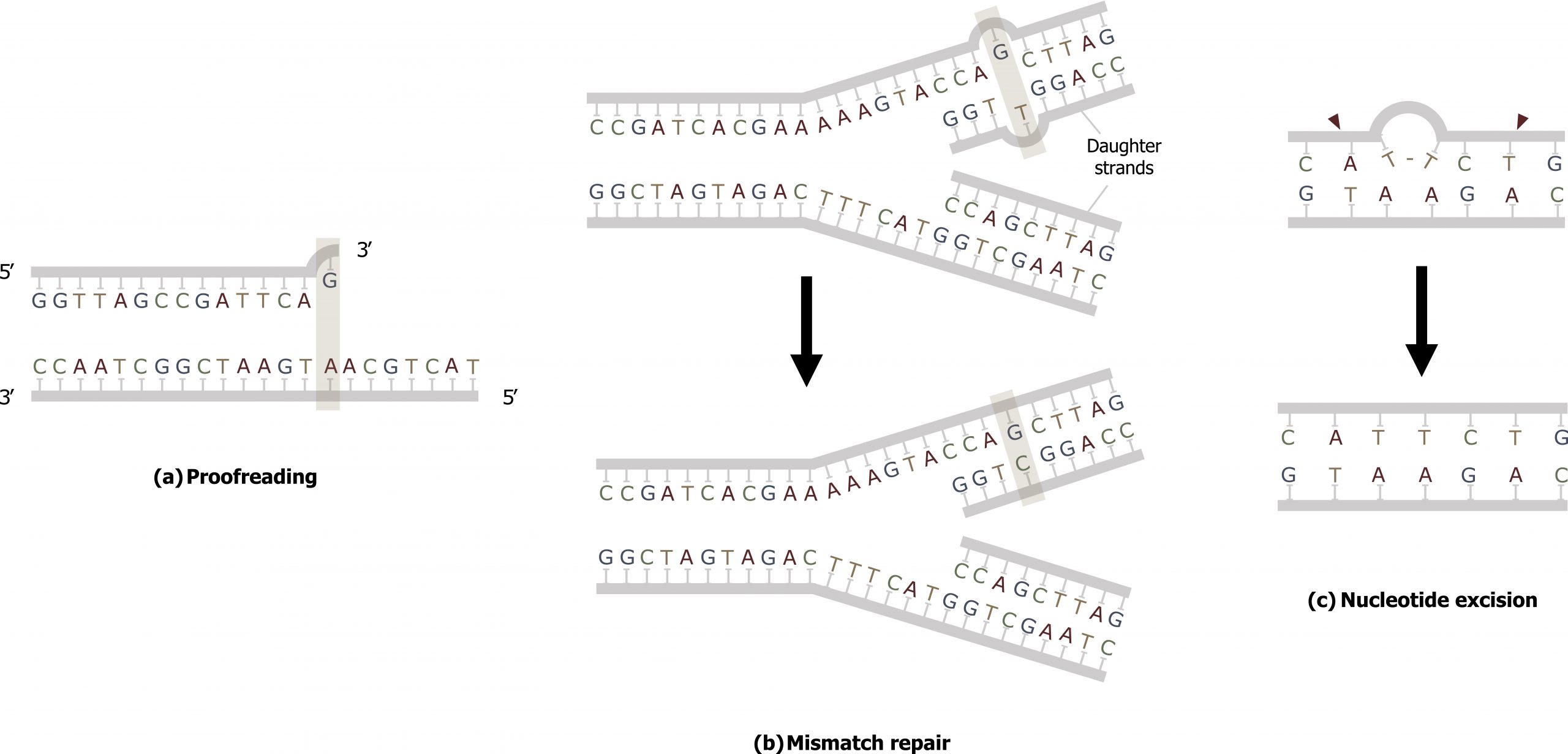
Most of the mistakes during DNA replication are promptly corrected by the proofreading ability of DNA polymerase itself. In proofreading, the DNA pol reads the newly added base before adding the next one, so a correction can be made. The polymerase checks whether the newly added base has paired correctly with the base in the template strand. If it is the right base, the next nucleotide is added. If an incorrect base has been added, the enzyme makes a cut at the phosphodiester bond and releases the wrong nucleotide. This is performed by the 3′ exonuclease action of DNA pol. Once the incorrect nucleotide has been removed, it can be replaced by the correct one (figure 10.5(a)).
Mismatch repair
Errors not addressed during replication are repaired through the process of mismatch repair (figure 10.5(b)). Specific repair enzymes recognize the mispaired nucleotide and excise part of the strand that contains it; the excised region is then resynthesized — typically during S phase of the cell cycle — and the enzymes involved are those used for DNA replication. If the mismatch remains uncorrected, it may lead to more permanent damage when the mismatched DNA is replicated. Deficiencies in this repair process can result in Lynch syndrome, which is characteristic of nonpolyposis colorectal cancer.
In prokaryotes, the parental strand is determined by the methyl groups on adenine bases, while the newly synthesized strand lacks them. Thus, DNA polymerase is able to remove the wrongly incorporated bases from the newly synthesized, non-methylated strand.
In eukaryotes, the mechanism is not very well understood, but it is believed to involve recognition of unsealed nicks in the new strand, as well as a short-term continuing association of some of the replication proteins with the new daughter strand after replication has completed.
Errors during DNA replication are not the only reason why mutations arise in DNA. Mutations, variations in the nucleotide sequence of a genome, can also occur because of damage to DNA. Such mutations may be of two types: induced or spontaneous. Induced mutations are those that result from an exposure to chemicals, UV rays, X-rays, or some other environmental agent. Spontaneous mutations occur without any exposure to any environmental agent; they are a result of natural reactions taking place within the body.
Nucleotide excision repair (NER)
Another type of repair mechanism, nucleotide excision repair, is similar to mismatch repair, except that it is used to remove large, bulky damaged bases rather than mismatched ones. The repair enzymes replace abnormal, bulky, bases by making a cut on both the 3′ and 5′ ends of the damaged base. The segment of DNA is removed and replaced with the correctly paired nucleotides by the action of DNA pol. Once the bases are filled in, the remaining gap is sealed with a phosphodiester linkage catalyzed by DNA ligase (figure 10.5(c)).
This repair mechanism is often employed when UV exposure causes the formation of pyrimidine dimers (thymine dimers). When exposed to UV light, thymines lying next to each other can form thymine dimers. In normal cells, they are excised and replaced. Xeroderma pigmentosa is a condition in which thymine dimerization from exposure to UV light is not repaired.
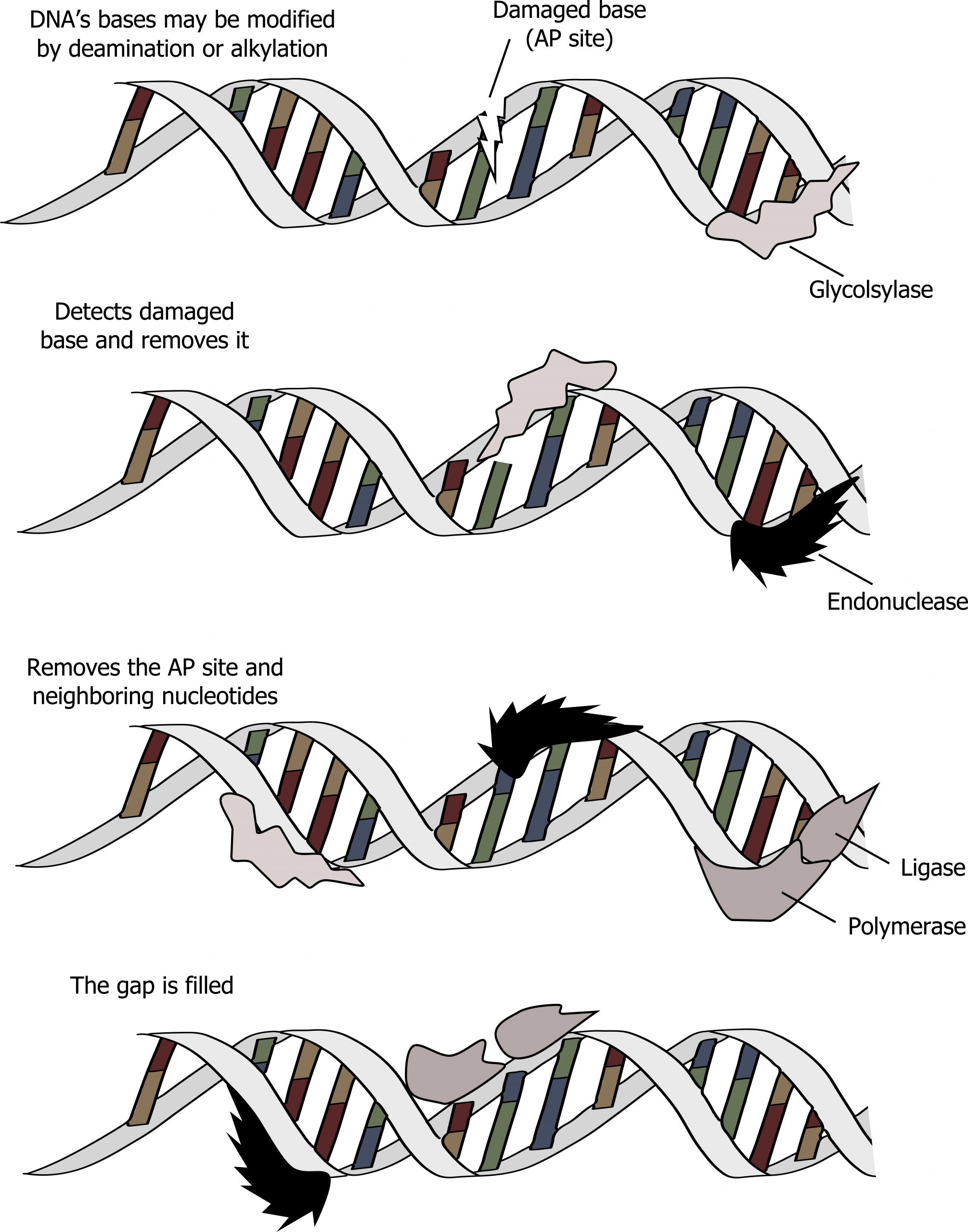
Base excision repair (BER)
The process of base excision repair (BER) is similar to NER but tends to repair small modifications to individual bases, such as deamination of cytosine to produce uracil. In this process, the aberrant base is detected by a glycosylase that will cleave the N-glycosidic bond joining the base to the deoxyribose sugar. This leaves an apurinic or apyrimidinic site (sugar phosphate backbone lacking a base), which is cleaved by an exonuclease and repaired through a similar process as mentioned above (figure 10.6).
Double-stranded break repair
Double-stranded breaks are caused by ionizing radiation, such as X-rays or radioactive particles. This can be repaired through two processes: nonhomologous end-joining and homologous recombination. The major difference between these two processes is in nonhomologous end-joining there is direct ligation of the two ends without the need for a DNA template. This can result in some DNA being lost in the process. In contrast, homologous recombination requires a DNA template to repair the break. This allows for restoration of the duplex without a loss of nucleotides.
10.2 References and resources
Text
Clark, M. A. Biology, 2nd ed. Houston, TX: OpenStax College, Rice University, 2018, Chapter 14: DNA Structure and Function.
Karp, G., and J. G. Patton. Cell and Molecular Biology: Concepts and Experiments, 7th ed. Hoboken, NJ: John Wiley, 2013, Chapter 10: The Nature of the Gene and the Genome, Chapter 12: The Cell Nucleus and the Control of Gene Expression, Chapter 13: DNA Replication.
Le, T., and V. Bhushan. First Aid for the USMLE Step 1, 29th ed. New York: McGraw Hill Education, 2018, 34, 38–40.
Nussbaum, R. L., R. R. McInnes, H. F. Willard, A. Hamosh, and M. W. Thompson. Thompson & Thompson Genetics in Medicine, 8th ed. Philadelphia: Saunders/Elsevier, 2016, Chapter 2: The Introduction to the Human Genome.
Figures
Grey, Kindred, Figure 10.5 Comparison on three types of repair. a) Proofreading b) Mismatch and c) Nucleotide excision repair. 2021. CC BY 4.0.
Grey, Kindred, Figure 10.6 Summary of Base excision repair. This is a similar process to NER but requires a glycosylase. 2021. https://archive.org/details/10.6_20210926. CC BY-SA 4.0. Added DNA double helix grooves by Biochemlife. CC BY SA 4.0. From Wikimedia Commons.
10.3 DNA Replication
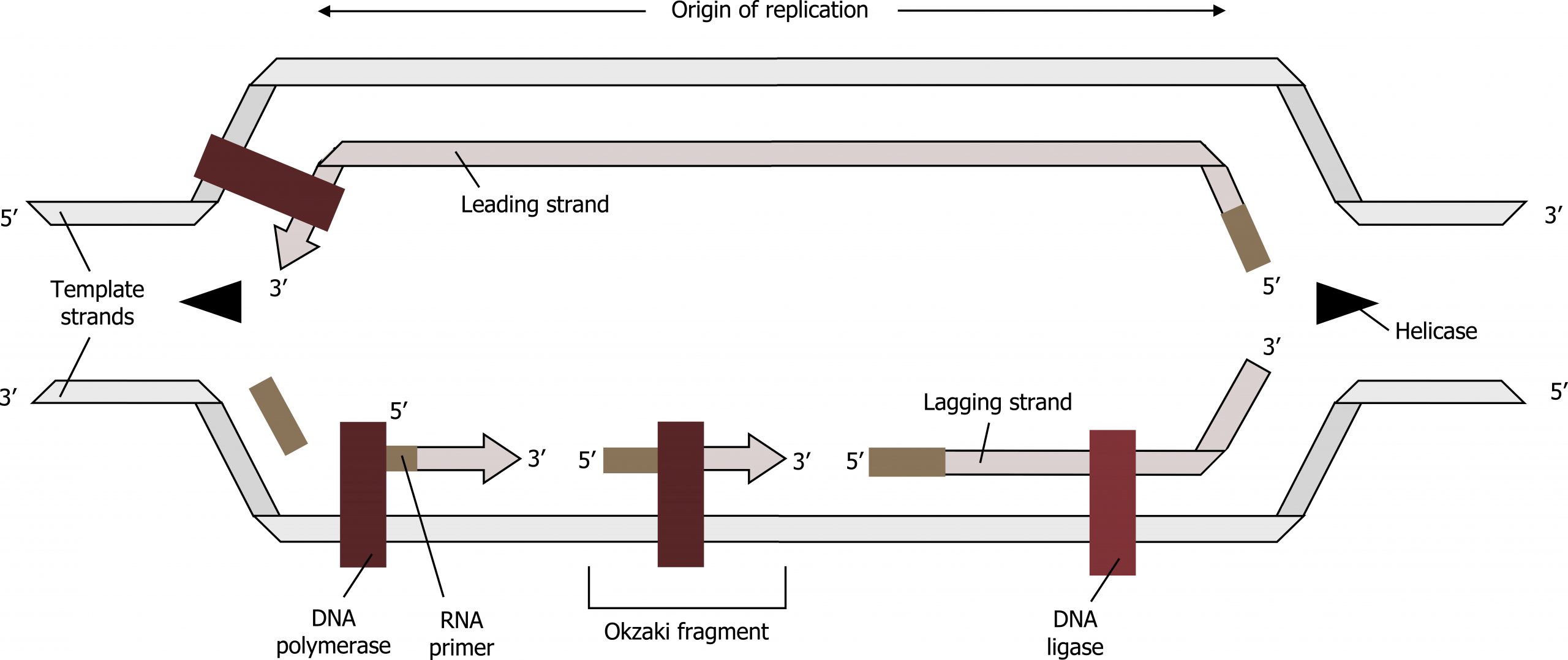
The process of DNA replication can be summarized as follows:
- DNA unwinds at the origin of replication.
- Helicase opens up the DNA-forming replication forks; these are extended bidirectionally.
- Single-strand binding proteins coat the DNA around the replication fork to prevent rewinding of the DNA.
- Topoisomerase binds at the region ahead of the replication fork to prevent supercoiling.
- Primase synthesizes RNA primers complementary to the DNA strand.
- DNA polymerase III starts adding nucleotides to the 3′-OH end of the primer.
- Elongation of both the lagging and the leading strand continues.
- RNA primers are removed by exonuclease activity.
- Gaps are filled by DNA pol I by adding dNTPs.
- The gap between the two DNA fragments is sealed by DNA ligase, which helps in the formation of phosphodiester bonds.
DNA replication
The essential steps of replication are the same for both prokaryotes and eukaryotes. Before replication can start, the DNA has to be made available as a template. Eukaryotic DNA is bound to basic proteins known as histones to form structures called nucleosomes. Histones must be removed and then replaced during the replication process, which helps account for the lower replication rate in eukaryotes. The chromatin (the complex between DNA and proteins) may undergo some chemical modifications, so that the DNA may be able to slide off the proteins or be accessible to the enzymes of the DNA replication machinery.
One of the key players in DNA replication is the enzyme DNA polymerase, also known as DNA pol, which adds nucleotides one-by-one to the growing DNA chain that is complementary to the template strand.
In prokaryotes, three main types of polymerases are known: DNA pol I, DNA pol II, and DNA pol III.
In eukaryotes there are fourteen are known polymerases, of which five are known to have major roles during replication and have been well studied. They are known as pol α, pol β, pol γ, pol δ, and pol ε.
How does the replication machinery know where to begin?
There are specific nucleotide sequences called origins of replication where replication begins. In prokaryotes, there is typically a single origin of replication on its one chromosome, and this is in contrast to eukaryotes that have many origins of replication across the chromosomes.
The origin of replication is recognized by certain proteins that bind to this site. An enzyme called helicase unwinds the DNA by breaking the hydrogen bonds between the nitrogenous base pairs. ATP hydrolysis is required for this process. As the DNA opens up, Y-shaped structures called replication forks are formed. Two replication forks are formed at the origin of replication, and these get extended bidirectionally as replication continues. Single-strand binding proteins coat the single strands of DNA near the replication fork to prevent the single-stranded DNA from winding back into a double helix.
DNA polymerase has two important restrictions. First, it is able to add nucleotides only in the 5′ to 3′ direction (a new DNA strand can be only extended in this direction). Second, it also requires a free 3′-OH group to which it can add nucleotides by forming a phosphodiester bond between the 3′-OH end and the 5′ phosphate of the next nucleotide. This essentially means that it cannot add nucleotides if a free 3′-OH group is not available.
Then how does it add the first nucleotide? The problem is solved with the help of a primer that provides the free 3′-OH end. RNA primase synthesizes an RNA segment that is about five to ten nucleotides long and complementary to the template DNA. Because this sequence primes the DNA synthesis, it is appropriately called the primer. DNA polymerase can now extend this RNA primer, adding nucleotides one-by-one that are complementary to the template strand.
The DNA tends to become more highly coiled ahead of the replication fork. Topoisomerase breaks and reforms DNAʼs phosphate backbone ahead of the replication fork, thereby relieving the pressure that results from this “supercoiling.” Single-strand binding proteins bind to the single-stranded DNA to prevent the helix from re-forming.
Because DNA polymerase can only extend in the 5′ to 3′ direction, and because the DNA double helix is antiparallel, there is a problem at the replication fork. The two template DNA strands have opposing orientations: one strand is in the 5′ to 3′ direction, and the other is oriented in the 3′ to 5′ direction. Only one new DNA strand, the one that is complementary to the 3′ to 5′ parental DNA strand, can be synthesized continuously toward the replication fork. This continuously synthesized strand is known as the leading strand. The other strand, complementary to the 5′ to 3′ parental DNA, is extended away from the replication fork in small fragments known as Okazaki fragments, each requiring a primer to start the synthesis. New primer segments are laid down in the direction of the replication fork, but each pointing away from it.
The overall direction of the lagging strand will be 3′ to 5′, and that of the leading strand 5′ to 3′. A protein called the sliding clamp holds the DNA polymerase in place as it continues to add nucleotides. The sliding clamp is a ring-shaped protein that binds to the DNA and holds the polymerase in place. As synthesis continues, the RNA primers are removed by the exonuclease activity of DNA pol I, which uses DNA behind the RNA as its own primer and fills in the gaps left by removal of the RNA nucleotides by the addition of DNA nucleotides. The nicks that remain between the newly synthesized DNA (that replaced the RNA primer) and the previously synthesized DNA are sealed by the enzyme DNA ligase, which catalyzes the formation of phosphodiester linkages between the 3′-OH end of one nucleotide and the 5′ phosphate end of the other fragment.
Once the chromosome has been completely replicated, the two DNA copies move into two different cells during cell division.
Telomere replication
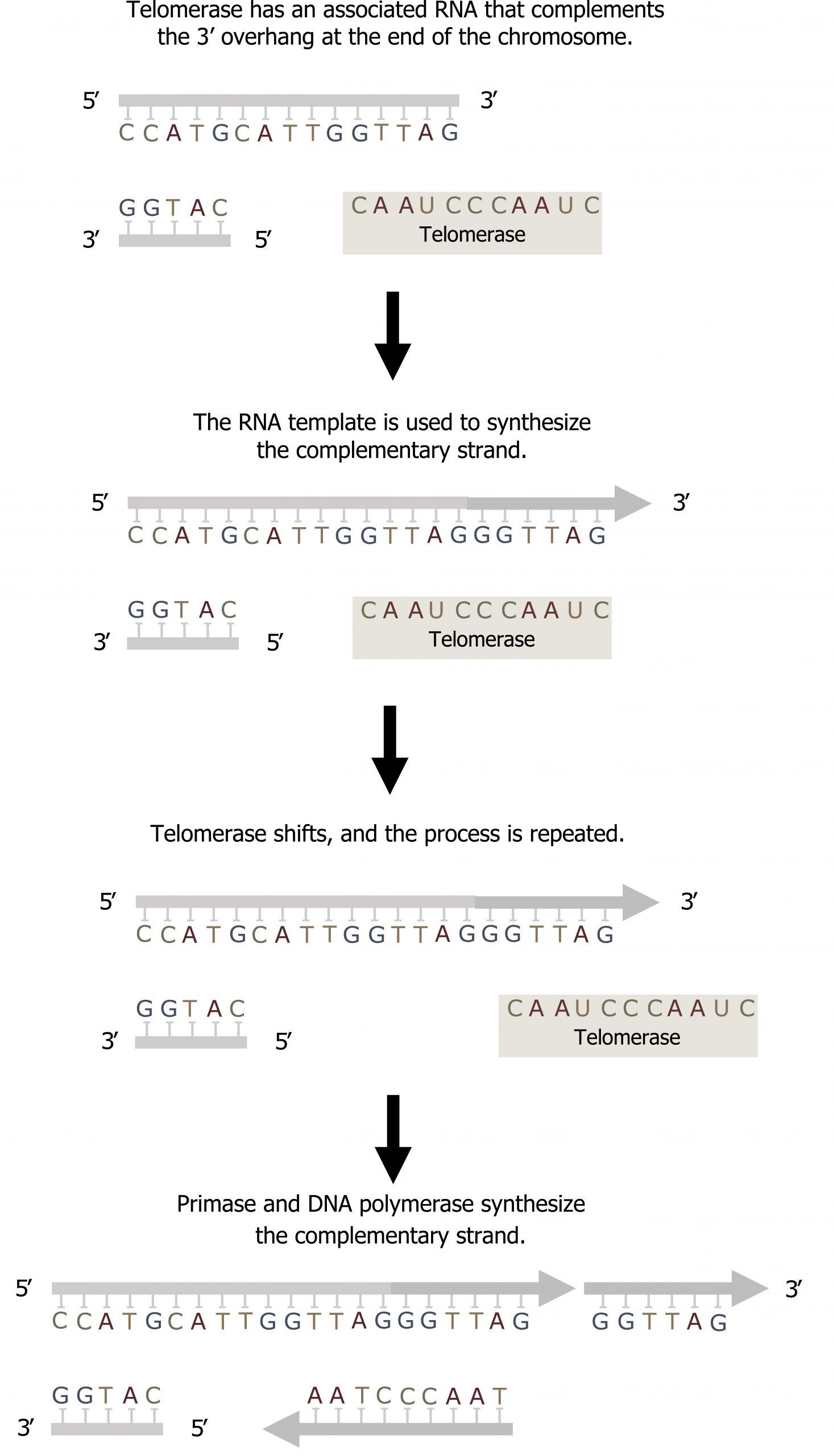
In eukaryotes, leading strand synthesis continues until the end of the chromosome is reached. On the lagging strand, DNA is synthesized in short stretches, each of which is initiated by a separate primer. When the replication fork reaches the end of the linear chromosome, there is no way to replace the primer on the 5ʼ end of the lagging strand.
The DNA at the ends of the chromosome thus remains unpaired, and over time these ends, called telomeres, may get progressively shorter as cells continue to divide.
Telomeres comprise repetitive sequences that code for no particular gene. In humans, a six-base-pair sequence, TTAGGG, is repeated 100 to 1,000 times in the telomere regions. In a way, these telomeres protect the genes from getting deleted as cells continue to divide. The telomeres are added to the ends of chromosomes by a separate enzyme, telomerase (figure 10.8), whose discovery helped in the understanding of how these repetitive chromosome ends are maintained. The telomerase enzyme contains a catalytic part and a built-in RNA template. It attaches to the end of the chromosome, and DNA nucleotides complementary to the RNA template are added on the 3′ end of the DNA strand. Once the 3′ end of the lagging strand template is sufficiently elongated, DNA polymerase can add the nucleotides complementary to the ends of the chromosomes. Thus, the ends of the chromosomes are replicated.
| Prokaryotic/Eukaryotic protein | Specific function |
|---|---|
| DNA pol I | Removes RNA primer and replaces it with newly synthesized DNA |
| DNA pol III/Pol δ and ε | Main enzyme that adds nucleotides in the 5′-3′ direction |
| Helicase | Opens the DNA helix by breaking hydrogen bonds between the nitrogenous bases |
| Ligase | Seals the gaps between the Okazaki fragments to create one continuous DNA strand |
| Primase/Pol α | Synthesizes RNA primers needed to start replication |
| Sliding clamp | Helps to hold the DNA polymerase in place when nucleotides are being added |
| Topoisomerase | Helps relieve the strain on DNA when unwinding by causing breaks, and then resealing the DNA |
| Single-strand binding proteins (SSB) | Binds to single-stranded DNA to prevent DNA from rewinding back |
Table 10.1: Prokaryotic DNA replication: enzymes and their function.
| Property | Prokaryotes | Eukaryotes |
|---|---|---|
| Origin of replication | Single | Multiple |
| Rate of replication | 1,000 nucleotides/s | 50 to 100 nucleotides/s |
| DNA polymerase types | 5 | 14 |
| Telomerase | Not present | Present |
| RNA primer removal | DNA pol I | RNase H |
| Strand elongation | DNA pol III | Pol α, pol δ, pol ε |
| Sliding clamp | Sliding clamp | PCNA |
Table 10.2: Difference between prokaryotic and eukaryotic replication.
10.3 References and resources
Text
Clark, M. A. Biology, 2nd ed. Houston, TX: OpenStax College, Rice University, 2018, Chapter 14: DNA Structure and Function.
Karp, G., and J. G. Patton. Cell and Molecular Biology: Concepts and Experiments, 7th ed. Hoboken, NJ: John Wiley, 2013, Chapter 10: The Nature of the Gene and the Genome, Chapter 12: The Cell Nucleus and the Control of Gene Expression, Chapter 13: DNA Replication.
Le, T., and V. Bhushan. First Aid for the USMLE Step 1, 29th ed. New York: McGraw Hill Education, 2018, 34, 38–40.
Nussbaum, R. L., R. R. McInnes, H. F. Willard, A. Hamosh, and M. W. Thompson. Thompson & Thompson Genetics in Medicine, 8th ed. Philadelphia: Saunders/Elsevier, 2016, Chapter 2: The Introduction to the Human Genome.
Figures
Grey, Kindred, Figure 10.7 Summary of DNA replication. 2021. https://archive.org/details/10.7_20210926. CC BY 4.0.
Grey, Kindred, Figure 10.8 Summary of Telomerase activity to fill the overhand on the lagging strand. 2021. CC BY 4.0.

