Appendix A: Bacteriology Resource
| Test | Visual | Notes |
|---|---|---|
| MacConkey (MAC) Media | 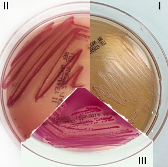 |
MacConkey Media: Bile salts and crystal violet are added to inhibit gram-positives; the fermentation of lactose produces acidic products that turn the crystal violet (also a pH indicator) pink. |
| Eosin Methylene Blue (EMB) Media | 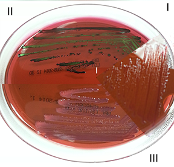 |
EMB Media: Eosin blue and methylene blue inhibits the growth of gram-positives; the fermentation of lactose produces acidic products that turn the methylene blue to purple or creates a green metabolic sheen with high lactose fermentation. |
| Kligler’s Iron Agar (KIA) | 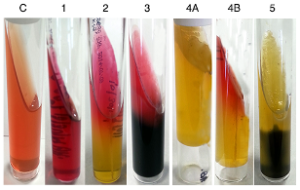 |
Contents of media: (C) control, (1) glucose, (2) lactose, (3) peptone, (4A) phenol red, (4B) sodium thiosulfate, and (5) iron
All Enterobacteriaceae will initially ferment glucose, changing the color from red to yellow. If the microorganism can ferment lactose, it will be fermented second, also producing acid end products and the media will stay yellow. If lactose cannot be fermented, the peptone will be utilized and basic products will be produced, changing the slant back to red. The black color is ferrous sulfide produced from the reaction of H2S with the iron in the media. |
| Urease Test |  |
If a microorganism produces urease, it can break-down urea in the media.
Ammonia is an alkaline product of this enzymatic reaction and changes the color of the pH indicator (phenol red) to pink. |
| Citrate Test |  |
If a microorganism produces citrase, citrate in the media is broken-down to oxaloacetic acid and acetic acid.
The oxaloacetic acid is further oxidized to pyruvic acid and CO2. The CO2 combines with Na in the media to form sodium bicarbonate. This is an alkaline product that changes the pH indicator (bromophenol blue) from green to blue. |
| SIM Test |  |
S-H2S production: similar process to KIA (above).
I-Indole production: tryptophan is broken down to ammonia, pyruvic acid, and indole. The indole reacts with Kovac’s reagent to produce a red color. M-motility: motility is observed as growth radiating outward along the length of the stab line. |
| Oxidase Test |  |
If the microorganism has cytochrome c then they oxidize the TMPD to a dark purple.
Microorganisms that lack cytochrome c can still use oxygen as a terminal electron acceptor but simply do not use cytochrome c. |
| Phenylethyl alcohol (PEA) agar | 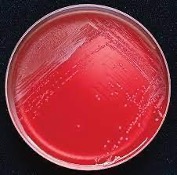 |
Allows the growth of gram-positive organisms, particularly cocci
Inhibits most gram-negative bacteria and fungi PEA agar is used to inhibit common contaminants such as Escherichia coli and Proteus species. |
| Tryptic Soy Agar (TSA) | 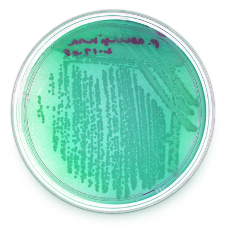 |
Allows the growth of gram-positive organisms, particularly cocci
Inhibits most gram-negative bacteria and fungi PEA agar is used to inhibit common contaminants such as Escherichia coli and Proteus species. |
| Mannitol salt agar or MSA | 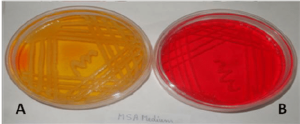 |
Mannitol salt agar or MSA contains a high concentration (about 7.5–10%) of salt (NaCl) which is inhibitory to most bacteria - making MSA selective against most Gram-negative and selective for some Gram-positive bacteria (Staphylococcus, Enterococcus and Micrococcaceae) that tolerate high salt concentrations. |
| Blood Agar Plate (BAP) | 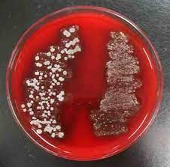 |
BAP tests the ability of an organism to produce hemolysins.
The degree of hemolysis by these hemolysins is helpful in differentiating members of the genera Staphylococcus, Streptococcus and Enterococcus. |
Table A.1: Biochemical tests and media used to differentiate Enterobacteriaceae
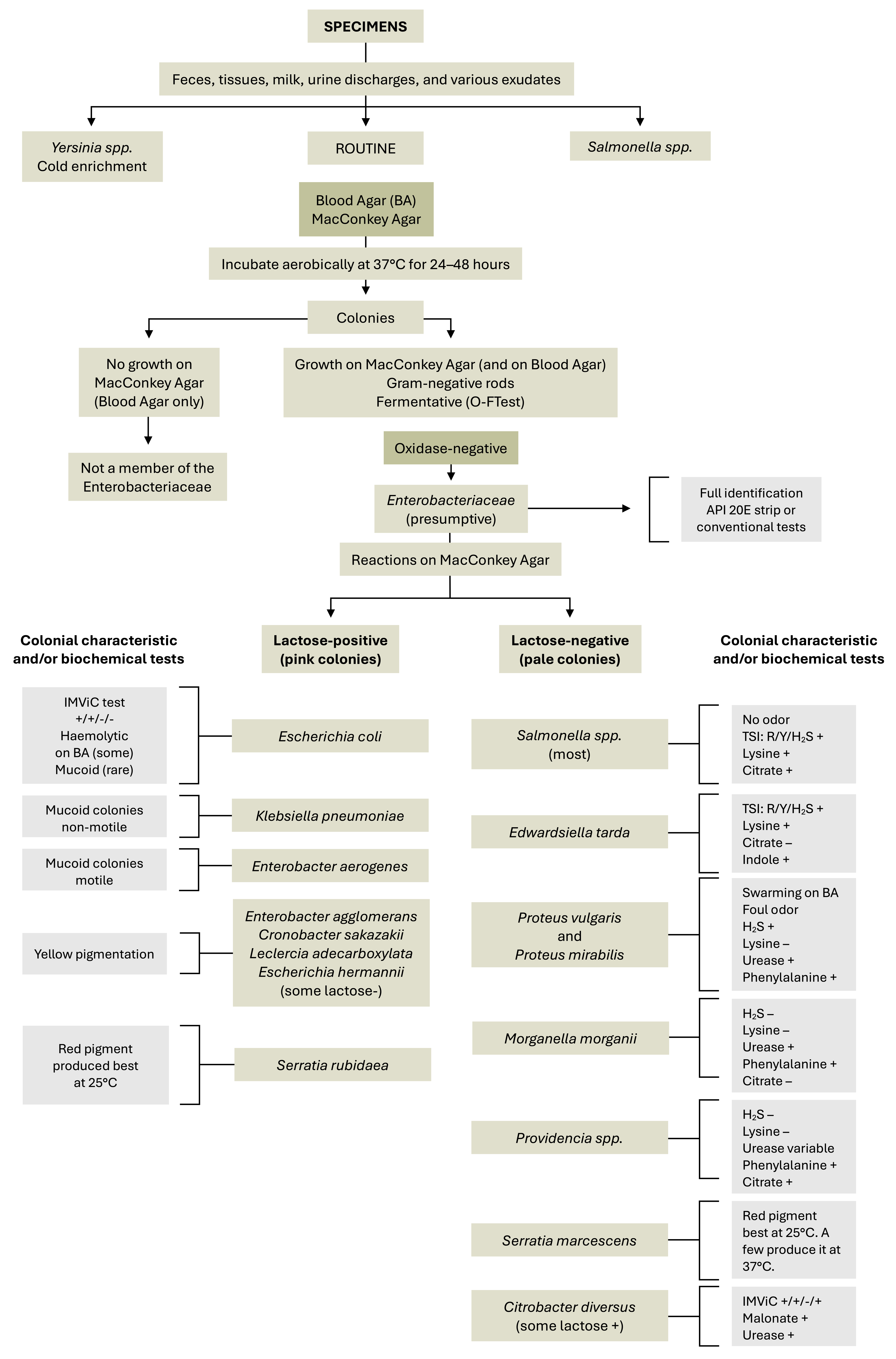
Figure Description
Figure A.1: Flow chart. Specimens. Points to feces, tissues, milk, urine discharges, and various exudates. Points to (1) yersinia spp. cold enrichment, (2) routine, and (3) salmonella spp. Blood agar (BA) / MacConkey Agar points to incubate aerobically at 37°C for 24–48 hours. Points to colonies. Points to (1) no growth on MacConkey Agar (blood agar only) / not a member of the enterobacteriaceae, and (2) growth on MacConkey Agar and on Blood Agar, gram-negative rods, fermentative (O-FTest). Oxidase negative. Points to enterobacteriaceae (presumptive). Points to full identification, API 20E strip or conventional tests. Also points to reactions on MacConkey Agar, which can be either lactose-positive (pink colonies) or lactose-negative (pale colonies). Colonial characteristics and/or biochemical tests of lactose positive: (1) Esterichia coli – IMViC test +/+/-/-, haemolytic on BA (some), mucoid (rare), (2) Klebsiella pneumoniae – Mucoid colonies non-motile, (3) Enterobacter aerogenes – Mucoid colonies motile, (4) Enterobacter agglomerans, Cronbacter sakazakii, Leclercia adecarboxylata, Escherichia hermannii (some lactose-) – yellow pigmentation, (5) Serratia rubidaea – red pigment produced best at 25°C. Colonial characteristics and/or biochemical tests of lactose negative: (1) Salmonella spp. (most) – no odor, TSI R/Y/H2S +, lysine +, citrate +, (2) Edwardsiella tarda – TSI R/Y/H2S +, lysine +, citrate -, indole +, (3) Proteus vulgaris and proteus mirabilis – swarming on BA, foul odor, H2S +, lysine -, urease +, phenylalanine +, (4) Morganella morganii – H2S -, lysine -, urease +, phenylalanine +, citrate -, (5) Providencia spp. – H2S -, lysine -, urease variable, phenylalanine +, citrate +, (6) Serratia marcescens – red pigment best at 25°C, a few produce it at 37°C, (7) Citrobacter diversus (some lactose +) – IMViC +/+/-/+, malonate +, urease +.
Figure Reference
Figure A.1: Specimens. Under fair use and redrawn. Visit https://veteriankey.com/enterobacteriaceae to access the figure, resources, and other information.

